If you register with LinkedIn, suggestions for reference to people you recognize are frequently presented, either because you might have worked at the identical university as you went in the identical company or in the identical industry.
However, the suggestions can sometimes surprise us, e.g. In view of the shortage of lack of skilled overlaps, you may ask yourself how LinkedIn could possibly learn about these real relationships.
The algorithms for artificial intelligence (AI) that drive these recommendations forward Graphics of Neural NetworkWhat is predicated on Graphics: Mathematical structures consisting of nodes and left (also known as “edges”), which they connect. For a social network like LinkedIn, a diagram might be generated by which the nodes represent every user, while the links are the connections between them.
These algorithms collect information from the immediate vicinity of each node – our direct connections to LinkedIn. Then aggregate this information and integrate into the unique node.
After this process, each profile reflects each its own data and that of its immediate network. This process might be carried out several times – within the second iteration, if we aggregate information from our neighbors, you have already got details about your personal neighbors, and consequently now we have information from the second neighborhood.
M. Hernaez / Biorererer.
A network of relationships
In these networks, it shouldn’t be only our own personal information that is vital, but additionally with which now we have connected and with whom our connections are connected. In The full version of Linkedins algorithmAs utilized in practice, there aren’t only nodes that represent people, but additionally other sorts of nodes corresponding to corporations or publications.
This signifies that the algorithm can receive information from our personal connections from each our personal connections and the content that now we have marked as favorites.
For example, if someone has their sister as a connection and “liked” contributions that their brother -in -law also likes, the algorithm can find that they not only share similar interests, but that they can also be personally connected.
Social -Media -Algorithms in Biomedicine
The development of a drug from scratch is incredibly expensive and time -consuming. The discovery process often resembles a funnel. All potential candidates come at the highest and after they’ve been limited in various research stages, just one stays in clinical studies. This drug will then (hopefully) be available for clinical use in the final population.
Although that is crucial, the complexity of this profile means Drug change has develop into increasingly common in recent many years. The aim of this process shouldn’t be to design latest medication, but to seek out latest uses for existing ones.
In order to treat an illness, we generally deal with the proteins chargeable for this. There are public and well -documented databases that contain details about which proteins aim for every medicine, and these databases have grown considerably in recent times.
One of probably the most steadily used databases, Drug benchwent from 841 approved medication when it was Published for the primary time published in 2006until 2,751 in his Last 2024 update. This growing availability of knowledge enables the usage of more complex models.
With this data volume we will create a graph network by which the nodes are medication and proteins and the links between them, as recorded in databases. As soon as now we have the network, we will use similar algorithms as in social media: For every medication, we add biochemical information in regards to the proteins with which it interacts through the known connections.
With this information, the model can then communicate the likelihood of a drug-protein interaction that we didn’t previously had within the database, because the algorithms can efficiently analyze large information volumes. These interactions can then be validated under laboratory conditions to save lots of money and time from the lengthy discovery process.
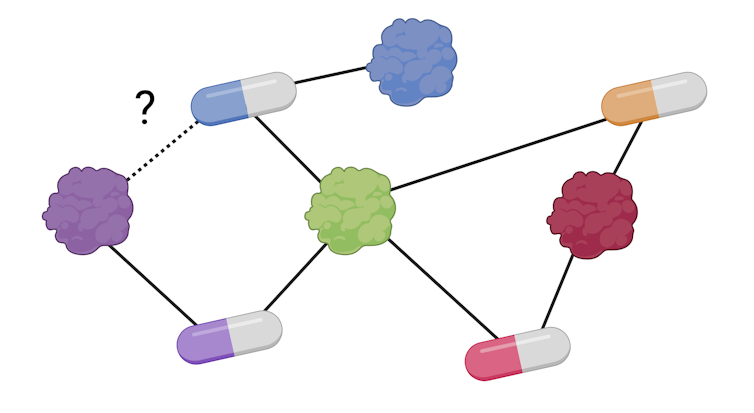
M. Hernáez / Biorender
Our research
In the laboratory for computer biology and translational genomics of the University of Navarra, we followed this concept to develop us GennA model that goals to construct a network between drugs and proteins. The implementation has already improved existing models, especially with regard to the term: We can evaluate around 23,000 interactions in only one minute.
While the model has good prediction functions, there continues to be room for improvement. For example, there are challenges within the evaluation of possible interactions with molecules that aren’t a part of the network or for which we only have a number of original data. Although it’s technically possible to generate an output, the model often gives little trust in these cases.
By overcoming these obstacles and with further research, these models could turn into systems in the longer term that give personalized recommendations for every patient.

